| |
المؤلفون / Authors
الملخص / Abstract
الكلمات المفتاحية / Keywords
أقسام الملف
Introduction
related work
Proposed Method
Possible Improvements
References
|
| Comparing and Improving Common Image Segmentation Algorithms for Brain Tumor Detection and Size Estimation |
| |
| Saad Alkentar[1], Abdulkareem Assalem[2], Bassem Assahwa[3] |
| Saad.zgm@gmail.com, assalem1@gmail.com, bassem74@netcourrier.com |
| 1. Phd Student at Al Baath University 2. Professor at Al Baath University 3.Professor at HIAST and SVU |
| |
| |
| Abstract |
|
| The accurate detection of a brain tumor in MRI images is one of the most important medical image-processing tasks. Early detection of a brain tumor increases the patient-survivor ratio. The tumor detection phase can heavily affect the treatment plans and can influence the patient’s lifespan. Tumor area is usually selected by trained doctors with clinical experience. But this process is a long-time consumer and can vary in accuracy depending on the doctor's experience and on the MRI device and image quality. Here comes the importance of a fully automated image segmentation system that can overcome those challenges. DL is heavily used for medical image processing lately. The new ML models are producing state-of-the-art results in image segmentation. We concentrated our study on FCN, UNET, and Masked-RCNN algorithms, we start by comparing those algorithms to find the best for this task and continue to propose additional processing steps that increased the overlap with the infected area by 8% compared to the original algorithm results |
|
| |
|
| Keywords: Image segmentation, medical image processing, MRI, brain tumor, FCN, UNET, Masked-RCNN, K-Means |
|
| |
|
| |
|
| Introduction |
|
| Deep learning (DL) has a large effect on multiple scientific fields. We are concentrating our study on one of the most important image-processing tasks: medical image segmentation. Lately, we have seen many DL algorithms giving great results in medical image segmentation [1, 2, 3, 4]. Many algorithms were suggested for different types of medical imaging technology, including X-Ray, visible-light, and MRI images. There are also many medical imaging datasets, like BraTS[5, 6, 7], KiTs19[8] and Covid19-20[9, 10]. Many studies worked on improving the accuracy of image segmentation by changing the neural network structure, changing the depth like RestNet[11] or the width like ResNext[12], or changing the connections between layers like in DenseNet[13]. Those modifications improved the accuracy of the network, but increasing the network dimensions and the number of parameters had a negative effect on small tumor detection. Structural changes generally gave about 1-2% [17] improvements in accuracy. |
|
| There are also algorithms based on attention mechanism, those algorithms use information from multiple images variant in time, and that information improved the accuracy for detection. This mechanism was first introduced in Natural Language Processing (NLP) [20] applications, and lately in images processing [21] and medical image processing. |
|
| We will concentrate our study on Fully Convolutional Networks (FCN) [22], UNET [1, 2], and Masked-RCNN [23]. We will use images from BraTS 2022 for training, evaluating, and testing our models. To improve the performance of those networks we will suggest a new processing layer based on K-Means [24] |
|
| |
|
| We will talk briefly about the mentioned algorithms in the related work section, the third section of this study will introduce training parameters for those three algorithms, and the fourth section discuss training results. The suggested extra layer is presented in the fifth section and its results in the seventh section of this study |
|
| |
|
| Related Work |
|
| We define image classification as finding the image content types (human, car, airplane…) while detection is finding the object location in the image. So semantic segmentation is classifying each pixel (or group of pixels) in an image. This section gives a brief description of the used algorithms |
|
| |
|
| 1. FCN |
|
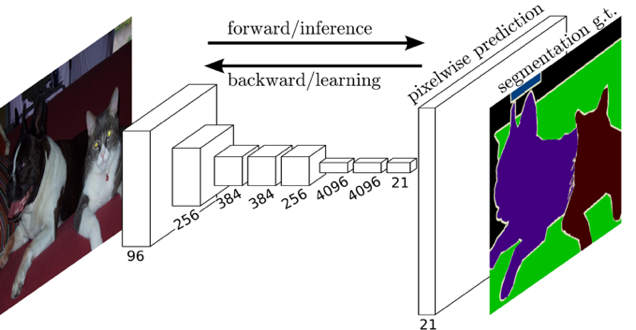
| FCN was one of the first Neural Networks (NN) used in semantic segmentation. by dropping the dense or fully connected layers in the network output, replacing them with convolutional layers (conv 1x1), and also recovering lost bits from conv filters by using up-pooling and up-conv layers, FCN output was in the same dimensions of its input. fig1 represents a simple FCN structure |
|
 |
|
| Fig 1 Simple FCN Architecture |
|
| FCN structures vary from a few simple layers structure to deeper networks like VGG and Resnet structures after making the previously mentioned modifications |
|
| |
|
| 2. UNET |
|
| UNET [1, 2] is considered one of the FCN structures, it divides the network into 2 blocks, the first one is called “The Encoder”, which uses filters to decrease the input image size similar to classical FCN, and the other block is called “The Decoder”, it has similar architecture to the encoder but in reverse, and it is used to recover the original image dimensions. The network is called U Net because of the shape of the network and seen in Fig 2 |
|
.png) |
|
| Fig 2 Simple UNET Architecture |
|
| UNET has many suggestions for encoder-decoder structures similar to those used in FCN [26, 27, 28] |
|
| |
|
| 3. Masked RCNN |
|
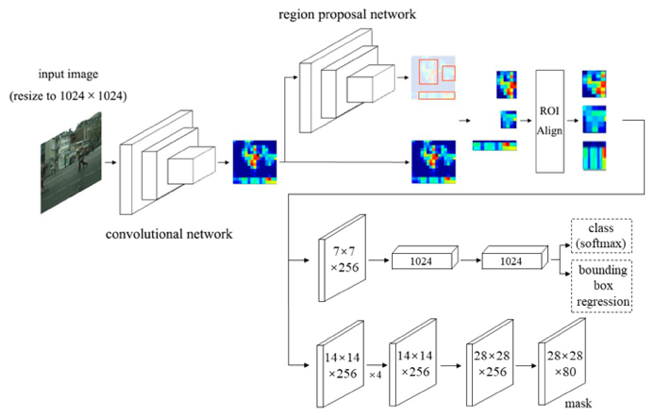
| Masked RCNN is considered the next version of Faster RCNN, it is two steps algorithm, the first step uses a Regions Proposal Network (RPN) to find regions with high target detection probability. The suggested regions (RPN output) are passed to FCN for segmentation. Masked RCNN can separate different objects from the same class due to its architecture, this is called instance segmentation. Fig 3 represents a simple Masked RCNN structure |
|
| |
|
| Comparison parameters |
|
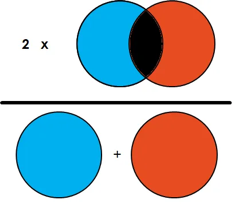
| We compared the mentioned algorithms based on image process time, procession (P), Recall (R), and Dice Coefficient (DC) for TP cases. DC can be calculated as shown in Fig 4 |
|
 |
|
| Fig 3 Masked RCNN Architecture |
|
 |
|
| Fig 4 Dice Coefficient |
|
| |
|
|
We count a True Positive (TP) case when there is an overlap between the predicted area (algorithm output) and the ground truth area, and we also calculate DC which represents the overlap area as Fig 4 represents. We count a False Negative (FN) when the algorithm fails to predict an existing tumor area. A false positive is counted when the algorithm predicts a tumor that doesn’t really overlap with the ground truth area. We calculate precession, recall, and F1 factor using those equations

|
|
 |
|
| Training parameters |
|
| To make a fair comparison we used the same training parameters for the three algorithms as presented in Table 1, we used Resnet as the backbone architecture for the three algorithms. |
|
| We used transfer learning to speed up training and get a good result with a small training set. We converted BraTS nii images into traditional images and filtered images without tumor areas. We used 1700 images for training, used 300 images for evaluation 1133 images for testing. Used a small learning rate of 0.0005. |
|
.jpg) |
|
| Comparison results |
|
| Tables 2 and 3 present comparison results. Table 2 shows statistical results and DC for TP cases. Table 3 presents P, R, and F1 which reflects the recall, precession relationship. We notice the similarity between FCN and UNET results, which can be caused by the similarity of network structures. Masked RCNN processing time is much longer than FCN and UNET, this can be caused by using RPN. Masked RCNN has higher F1 compared to other algorithms because of RPN. In spite of the high detection accuracy, all algorithms suffer limited DC values. We notice that UNET has the higher DC among all |
|
| Proposed algorithm |
|
| We notice the high detection accuracy of the three algorithms, but they suffer from limited overlap area with the ground truth tumor area. To improve their performance, we added K-Means layer |
|
| |
|
| 1.We start by applying DL algorithm (like UNET) which predicts base tumor areas, this prediction is presented as mask |
|
| 2.We extract the area around the predicted tumor area by applying pyramid-up and pyramid-down to the mask, and extract the area selected by the new mask |
|
| 3.We applied K-Means on the images extracted from the previous step |
|
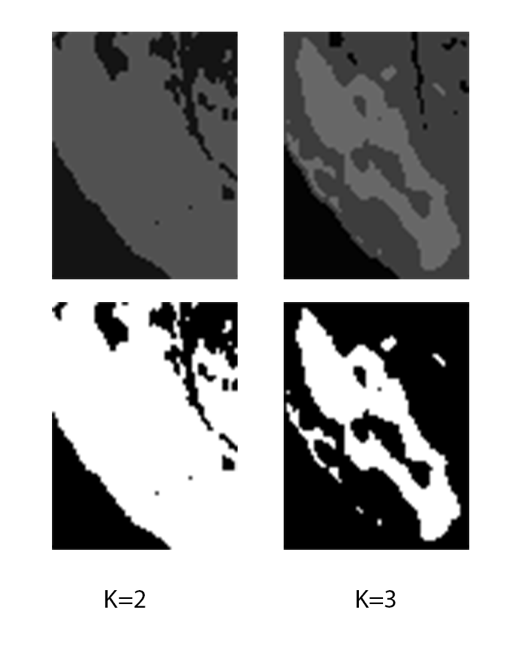
| 4.We choose k=2 when extracted are contains only tumor and healthy cells, we choose k=3 when the extracted area contains image background with tumor and healthy cells (Fig 6 shows a case of k=3) |
|
| 5.K-Means number of output masks equals the value of k, we choose the mask that selects the area with the greatest average mean |
|
.png) |
|
| Fig 5 Proposed algorithm |
|
 |
|
| Fig 6 Example of area with background |
|
| Fig 5 shows the proposed algorithm steps and Table 4 shows the results. We tested it with UNET and compared it with the latest studies. The suggested algorithm has an edge over the latest studies. This algorithm can be applied to any DL segmentation algorithm. The suggested algorithm doesn’t affect precession or recall, but it can greatly affect the intersection of predicted and ground truth areas. |
|
| While writing this study, we came across a similar paper called “Automatic Brain Tumor Segmentation Using Cascaded FCN with DenseCRF and K-Means” [25] |
|
.jpg) |
|
| The mentioned paper uses multiple layers of FCN, the first layer detects the whole tumor area and the second one finds the most infected area. The paper uses K-Means after the first FCN, then decide the pixel status (infected/ healthy) based on the scoring system between K-Means and the second layer FCN. The mentioned paper recorded a 1% improvement in DC. We can summarize the differences from our algorithm by those points |
|
| 1.Our algorithm modifies the DL algorithm mask to get the close neighborhood of the tumor area while the paper uses the proposed DL region without modification. |
|
| 2.Our algorithm discusses two values of K while the paper uses only one value k=2. |
|
| 3.Our algorithm uses the K-Means mask regardless of DL’s original mask, while the paper uses a scoring system between FCN and K-Means. |
|
|
4.Our algorithm gave an 8% improvement in DC while the paper’s algorithm gave a 1% DC improvement for whole tumor detection

|
|
| Possible improvements |
|
| We can use the proposed algorithm with any DL detection network, and it can be further improved by tuning K values or even mixing it with other algorithms as mentioned in [25]. We can also test the same approach with other traditional segmentation algorithms like super-pixel or GrapCut. |
|
| |
|
| Fig 7 presents samples of training results. The first row contains original images, the second row contains ground truth masks, the third row shows FCN results, UNET masks are shown in the fourth row and the fifth row shows Masked RCNN results |
|
| |
|
.png) |
|
| Fig 7 Samples of training results for the three algorithms |
|
| |
|
| References |
|
| 1.Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234- 241). Springer, Cham. |
|
| 2.Zhou, Z., Siddiquee, M. M. R., Tajbakhsh, N., & Liang, J. (2018). Unet++: A nested u-net architecture for medical image segmentation. In Deep learning in medical image analysis and multimodal learning for clinical decision support (pp. 3-11). Springer, Cham. 13 |
|
| 3.Jha, D., Smedsrud, P. H., Riegler, M. A., Johansen, D., De Lange, T., Halvorsen, P., & Johansen, H. D. (2019, December). Resunet++: An advanced architecture for medical image segmentation. In 2019 IEEE International Symposium on Multimedia (ISM) (pp. 225-2255). IEEE. |
|
| 4.Fan, D. P., Ji, G. P., Zhou, T., Chen, G., Fu, H., Shen, J., & Shao, L. (2020, October). Pranet: Parallel reverse attention network for polyp segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 263-273). Springer, Cham. |
|
| 5.Menze, B. H., Jakab, A., Bauer, S., Kalpathy-Cramer, J., Farahani, K., Kirby, J., ... & Van Leemput, K. (2014). The multimodal brain tumor image segmentation benchmark (BRATS). IEEE transactions on medical imaging, 34(10), 1993-2024. |
|
| 6.Bakas, S., Akbari, H., Sotiras, A., Bilello, M., Rozycki, M., Kirby, J. S., ... & Davatzikos, C. (2017). Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Scientific data, 4(1), 1-13. |
|
| 7.Bakas, S., Reyes, M., Jakab, A., Bauer, S., Rempfler, M., Crimi, A., ... & Jambawalikar, S. R. (2018). Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv preprint arXiv:1811.02629. |
|
| 8.Heller, N., Sathianathen, N., Kalapara, A., Walczak, E., Moore, K., Kaluzniak, H., ... & Weight, C. (2019). The kits19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. arXiv preprint arXiv:1904.00445. |
|
| 9.Roth, H., Xu, Z., Diez, C. T., Jacob, R. S., Zember, J., Molto, J., ... & Linguraru, M. (2021). Rapid Artificial Intelligence Solutions in a Pandemic-The COVID-19-20 Lung CT Lesion Segmentation Challenge. |
|
| 10.An, P., Xu, S., Harmon, S., Turkbey, E., Sanford, T., Amalou, A., ... & Wood, B. (2020). CT Images in Covid-19 [Data set]. The Cancer Imaging Archive. Mode of access: https://doi. org/10.7937/tcia. |
|
| 11.He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778). |
|
| 12.Xie, S., Girshick, R., Dollár, P., Tu, Z., & He, K. (2017). Aggregated residual transformations for deep neural networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1492-1500). |
|
| 13.Huang, G., Liu, Z., Van Der Maaten, L., & Weinberger, K. Q. (2017). Densely connected convolutional networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 4700-4708). |
|
| 14. Szegedy, C., Liu, W., Jia, Y., Sermanet, P., Reed, S., Anguelov, D., ... & Rabinovich, A. (2015). Going deeper with convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 1-9). |
|
| 15.Zhao, H., Shi, J., Qi, X., Wang, X., & Jia, J. (2017). Pyramid scene parsing network. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 2881-2890). |
|
| 16.Chen, L. C., Papandreou, G., Schroff, F., & Adam, H. (2017). Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587. |
|
| 17.Hesamian, M. H., Jia, W., He, X., & Kennedy, P. (2019). Deep learning techniques for medical image segmentation: achievements and challenges. Journal of digital imaging, 32(4), 582-596. |
|
| 18.Ngo, D. K., Tran, M. T., Kim, S. H., Yang, H. J., & Lee, G. S. (2020). Multi-task learning for small brain tumor segmentation from MRI. Applied Sciences, 10(21), 7790. |
|
| 19.Taghanaki, S. A., Abhishek, K., Cohen, J. P., Cohen-Adad, J., & Hamarneh, G. (2021). Deep semantic segmentation of natural and medical images: a review. Artificial Intelligence Review, 54(1), 137-178. |
|
| 20.Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., ... & Polosukhin, I. (2017). Attention is all you need. In Advances in neural information processing systems (pp. 5998-6008). |
|
| 21.Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., ... & Houlsby, N. (2020). An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929. |
|
| 22.Long, J., Shelhamer, E. and Darrell, T., 2015. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 3431-3440). |
|
| 23.He,K.,Gkioxari,G.,Dolla ́r,P.,andGirshick,R.,“Maskr-cnn,”in[ProceedingsoftheIEEEinternationalconference on computer vision], 2961–2969 (2017). |
|
| 24.Praveena, M. and Ms, S., 2010. Optimization fusion approach for image segmentation using k-means algorithm. International Journal of Computer Applications, 2(7). |
|
| 25.Lizhu Yang, Weinan Jiang, Hongkun Ji, Zijun Zhao, Xukang Zhu, Alin Hou. "Automatic Brain Tumor Segmentation Using Cascaded FCN with DenseCRF and K-means" 2019 IEEE/CIC International Conference on Communications in China |
|
| 26.Neil Micallef, Dylan Seychell, Claude J. Bajada; Exploring the U-Net++ Model for Automatic Brain Tumor Segmentation; IEEE 2021 |
|
| 27.Jianwei Lin, Jiatai Lin, Cheng Lu, Hao Chen, Huan Lin, Bingchao Zhao, Zhenwei Shi, Bingjiang Qiu, Xipeng Pan, Zeyan Xu, Biao Huang, Changhong Liang, Guoqiang Han, Zaiyi Liu, Chu Han; CKD-TransBTS: Clinical Knowledge-Driven Hybrid Transformer with Modality-Correlated Cross-Attention for Brain Tumor Segmentation; IEEE 2023 |
|
|
28.Yunjiao Deng, Yulei Hou, Jiangtao Yan, And Daxing Zeng; ELU-Net: An Efficient and Lightweight U-Net for Medical Image Segmentation. IEEE 2022

|
|
|